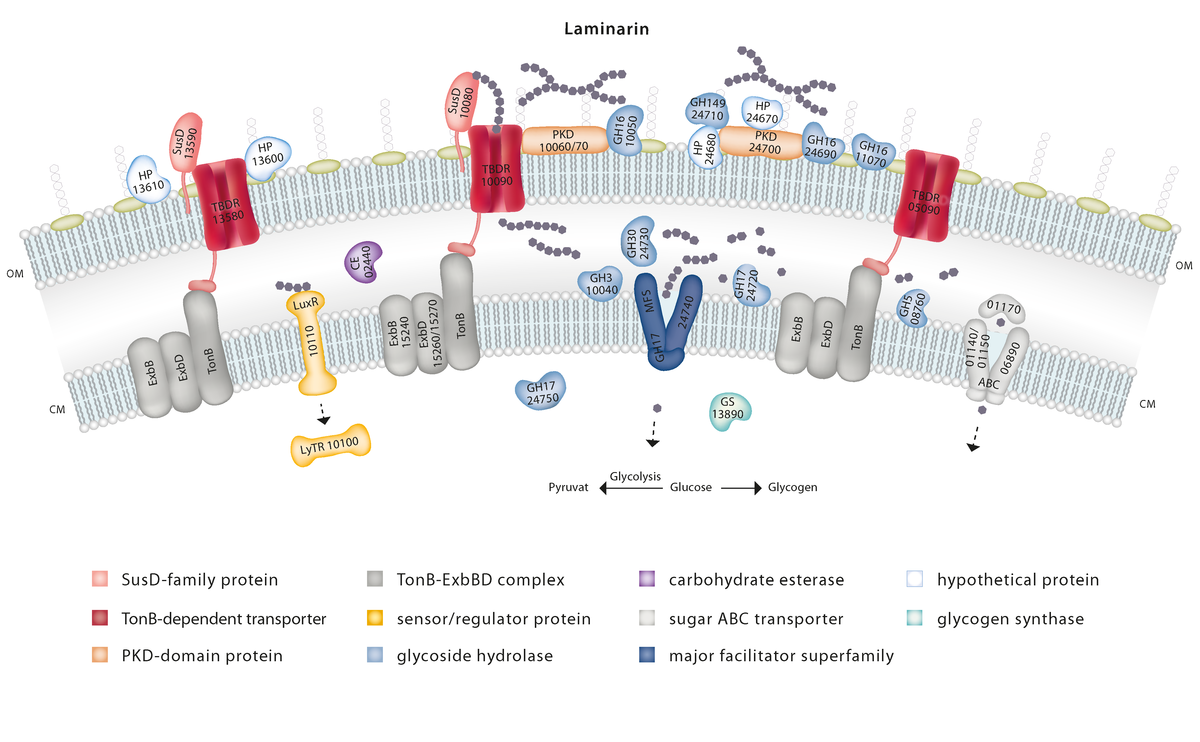
Molecular mechanisms of marine polysaccharide degradation
Algae blooms, i.e., massive proliferation of algae in marine ecosystems, are caused by seasonal changes in environmental conditions and nutrient contaminations, such as agricultural runoffs. These large quantities of algal biomass have detrimental impact on coastal ecosystems, tourism, and aquaculture. At the same time, they present a hitherto unrecognized sustainable resource of rare marine sugars. A complex range of nutrients released upon algae death presents fertile ground for bacteria. These nutrients include structurally versatile algal polysaccharides. To tap this partially recalcitrant food source, bacterial communities possess a diverse repertoire of highly specialized enzymes, substrate-binding proteins and transporters. Interestingly, individual bacteria pursue quite distinct strategies to prevail against nutritional competitors or to profit from each other. These foci are central to our DFG research unit POMPU (FOR 2406; www.pompu-project.de).
In cooperation with the Max Planck Institute for Marine Microbiology, Bremen, and the Alfred Wegener Institute (AWI), Helmholtz Centre for Polar and Marine Research, Bremerhaven, we investigate microalgae blooms at the long-term sampling site Helgoland Roads (Kabeltonne) near the island Helgoland in the German Bight (North Sea). Our studies revealed that massive multiplication of microalgae in the water column is accompanied by a subsequent increase in bacterial biomass. On closer inspection of the bacterial community, we observed on the one hand a specific succession of individual bacterial groups, and on the other hand the recurrence of individual strains within this pattern over several years of sampling. Among the bacterial groups that dominated algal blooms were representatives of the Flavobacteriia, which are capable of degrading a diverse range of algal polysaccharides. In the course of an algae bloom, bacteria preferably utilize easily digestible carbohydrates first, before they turn to structurally more complex algal polysaccharides. The respective genes that encode proteins required for polysaccharide depolymerization are often co-localized in "polysaccharide-utilization loci" (PULs) in the bacterial genome. Since not every bacterium can use the entire spectrum of polysaccharides, a wide variety of bacterial groups with distinct substrate specificities are required to cooperate to enable the concerted breakdown of total algal biomass. These consortia integrate bacteria with different growth and survival strategies, i.e. both, generalists with many PULs for degradation of various substrates, and specialists with only few specific PULs. Either strategy entails dedicated adaptations that merit further exploration.
To illuminate processes and interactions occurring during algal blooms, we use metagenomics and metaproteomics. These analyses comprise the core of our research focus. Another key aspect is the detailed examination of selected bacterial representatives isolated during algal bloom situations. We are specifically interested in functional characterization of PULs and degradation enzymes of these polysaccharide-utilizing bacteria. To decipher these molecular mechanisms of polysaccharide degradation, we apply proteome analyses as well as various molecular biological, analytical and microscopical methods. This enables us to identify enzymes that catalyze the targeted disassembly of polysaccharides into defined oligo- and monomers. These enzymes are tested for potential pharmaceutical and biotechnological applications. These studies are conducted in close cooperation with the Institutes of Microbiology and Biochemistry, University of Greifswald; and the Station Biologique de Roscoff, France.
Responsible staff members:
Dr. Marie-Katherin Zühlke, Dr. Stephanie Markert, Alexandra Dürwald, Irena Beidler, Daniel Bartosik
Literature:
- Kappelmann L, Krüger K, Hehemann JH, Harder J, Markert S, Unfried F, Becher D, Shapiro N, Schweder T, Amann RI, Teeling H. 2019. Polysaccharide utilization loci of North Sea Flavobacteriia as basis for using SusC/D-protein expression for predicting major phytoplankton glycans. ISME J. 13(1):76-91.
- Koch H, Dürwald A, Schweder T, Noriega-Ortega B, Vidal-Melgosa S, Hehemann JH, Dittmar T, Freese HM, Becher D, Simon M, Wietz M. 2019. Biphasic cellular adaptations and ecological implications of Alteromonas macleodii degrading a mixture of algal polysaccharides. ISME J. 13(1):92-103.
- Unfried F, Becker S, Robb CS, Hehemann JH, Markert S, Heiden SE, Hinzke T, Becher D, Reintjes G, Krüger K, Avcı B, Kappelmann L, Hahnke RL, Fischer T, Harder J, Teeling H, Fuchs B, Barbeyron T, Amann RI, Schweder T. 2018. Adaptive mechanisms that provide competitive advantages to marine bacteroidetes during microalgal blooms. ISME J. 12(12):2894-2906.
- Chen J, Robb CS, Unfried F, Kappelmann L, Markert S, Song T, Harder J, Avcı B, Becher D, Xie P, Amann RI, Hehemann JH, Schweder T, Teeling H. 2018. Alpha- and beta-mannan utilization by marine Bacteroidetes. Environ Microbiol. 20(11):4127-4140.
- Reisky L, Büchsenschütz HC, Engel J, Song T, Schweder T, Hehemann JH, Bornscheuer UT. 2018. Oxidative demethylation of algal carbohydrates by cytochrome P450 monooxygenases. Nat Chem Biol. 14(4):342-344.
- Hehemann JH, Truong LV, Unfried F, Welsch N, Kabisch J, Heiden SE, Junker S, Becher D, Thürmer A, Daniel R, Amann R, Schweder T. 2017. Aquatic adaptation of a laterally acquired pectin degradation pathway in marine Gammaproteobacteria. Environ Microbiol. 19(6):2320-2333.
- Kabisch A, Otto A, König S, Becher D, Albrecht D, Schüler M, Teeling H, Amann RI, Schweder T. 2014. Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes 'Gramella forsetii' KT0803. ISME J. 8(7):1492-502.
- Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, Kassabgy M, Huang S, Mann AJ, Waldmann J, Weber M, Klindworth A, Otto A, Lange J, Bernhardt J, Reinsch C, Hecker M, Peplies J, Bockelmann FD, Callies U, Gerdts G, Wichels A, Wiltshire KH, Glöckner FO, Schweder T, Amann R. 2012. Substrate-Controlled Succession of Marine Bacterioplankton Populations Induced by a Phytoplankton Bloom. Science. 336(6081):608-11.